Rational Design Identifies Chemical Modifications That Enhance Site-Directed RNA Editing
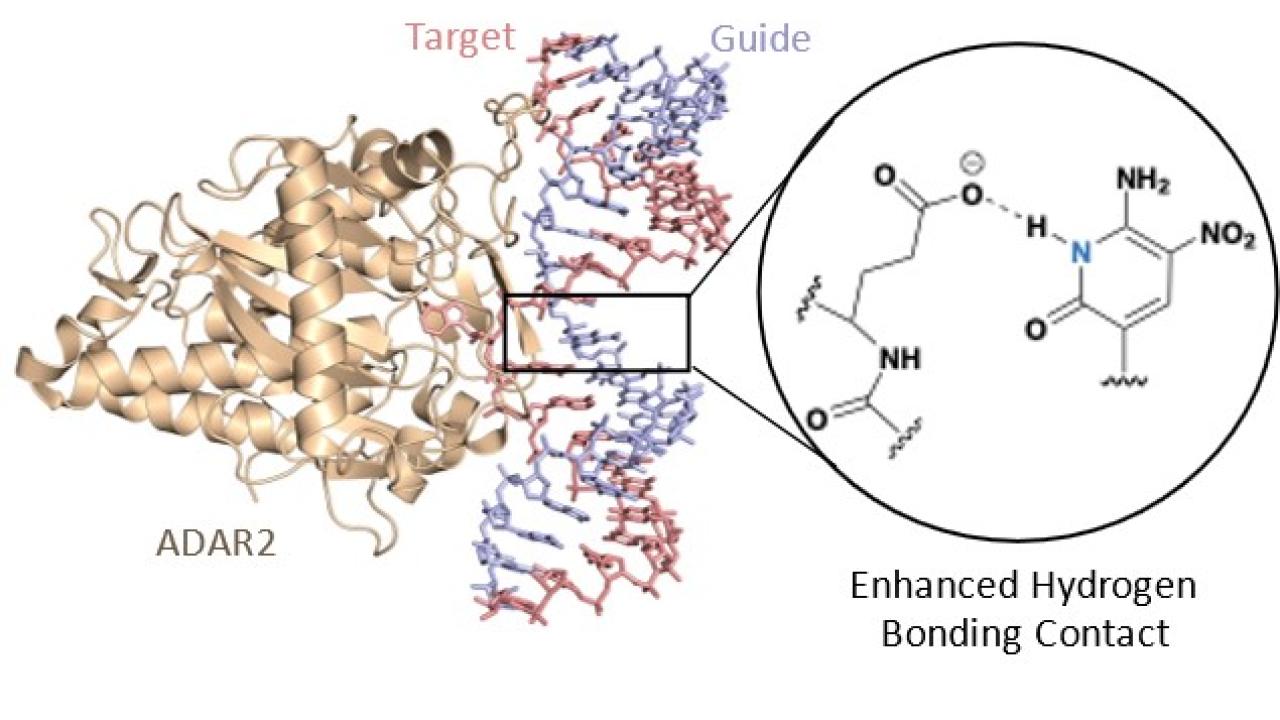
In a recent JACS publication, the Beal lab in collaboration with the Fisher lab, Tantillo lab, and ProQR Therapeutics, used structural information from enzyme-RNA complexes to design chemical modifications that increase RNA editing yields via Adenosine Deaminases Acting on RNA (ADARs). Enzymes that can site-specifically alter nucleic acids can be used to correct disease-causing mutations. Recruiting endogenous human enzymes for this purpose, such as the ADAR RNA editing enzymes, can minimize barriers associated with delivery and off-target reactions that are complications of other popular therapeutic editing strategies. Directed RNA editing requires the use of guide strands that can lead to high editing yields. Therefore, guide strand optimization is an active area of research. The nucleotide modification identified in this work led to a significant increase in editing yields and advances the approach of recruiting endogenous ADARs for site-directed RNA editing.
More information at acspubs.co/7QBP50EDGPb
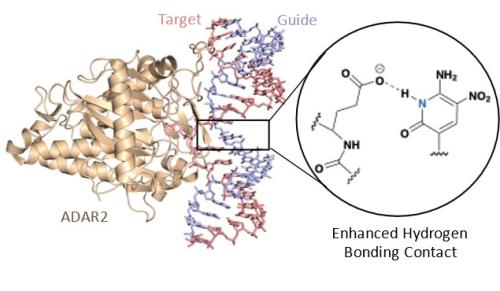
A new crystal structure (PDB ID: 7KFN) of ADAR2 bound to duplex RNA bearing a chemically modified guide strand.
