Detection of OG:A Lesion Mispairs by MutY Relies on a Single His Residue and the 2‑Amino Group of 8‑Oxoguanine
The chemical basis for detecting rare DNA lesions by base excision repair enzymes has captivated the imagination of the DNA repair field since it represents a statistically improbable feat. A collaboration between Sheila David’s lab and Andrea Lee’s lab at the University of Vermont provided key insights into the lesion recognition process by one such DNA repair enzyme. MutY, and it’s human homolog MUTYH, are adenine glycosylases proficient in locating rare and mutagenic OG:A mispairs and distinguishing them from structurally similar undamaged DNA.
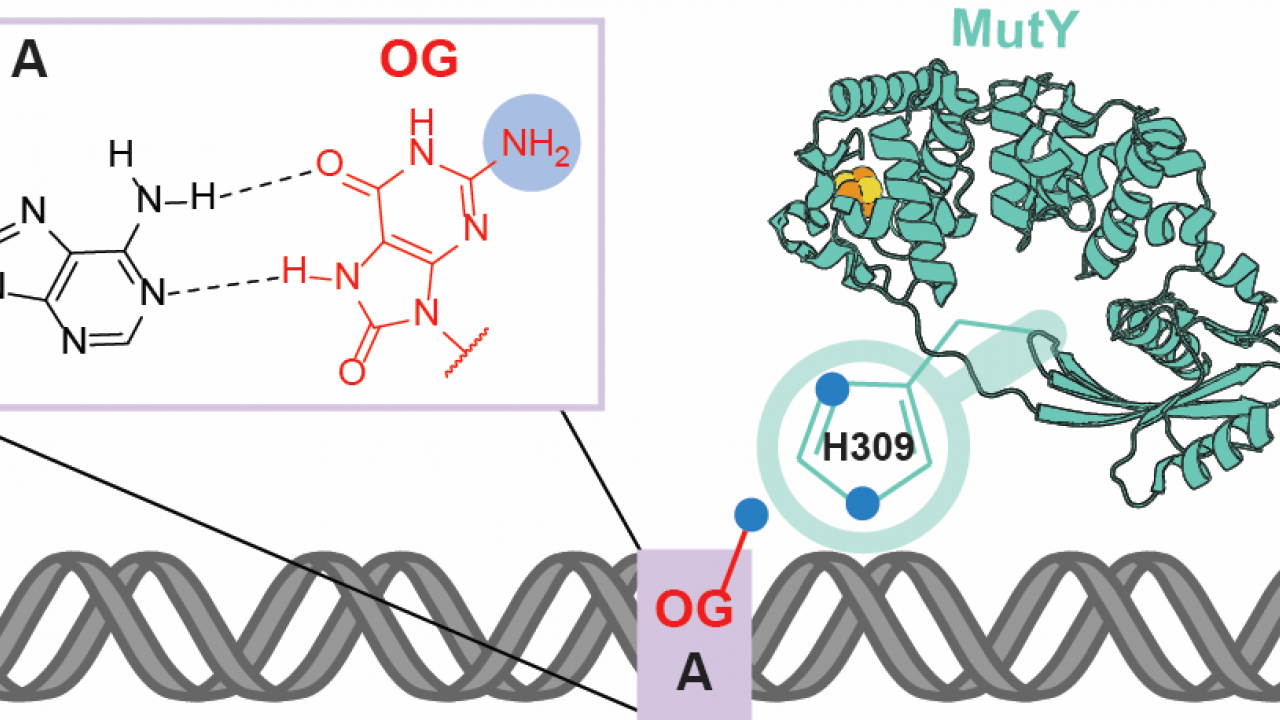
Through a combination of in vitro biochemical assays, cellular repair assays and single molecule visualization, a recent publication (Lee et. al. JACS 2020) reveals that MutY uses a conserved histidine residue to recognize the 2-amino group of OG positioned in the major groove of the DNA helix. Single molecule visualizations show that WT MutY traverses rapidly on undamaged DNA but exhibits long pauses on damaged sites on OG:A containing DNA. The absence of either the 2-amino group or the histidine residue caused the enzyme to bypass the damaged sites, indicating a lack of recognition. Interestingly, though the removal of either feature did not significantly affect the catalytic activity of the enzyme, overall cellular repair was completely disrupted, showing that successful repair is contingent on recognition of the damaged site.
Upon extrapolating the results to the human enzyme, these findings can be used to predict functional deficiencies in MUTYH and enable the development of chemical biology tools to target the enzyme.
Find out more at: https://pubs.acs.org/doi/10.1021/jacs.0c04284
